Overview
Metadata ingestion in Validatar allows for the automatic import of metadata from a data source. By harnessing the capabilities of metadata ingestion, users can benefit from automated test generation, data profiling, and a real-time overview of their dataset, including its changes over time. This feature supports both database-based and script-based data sources, providing flexibility in data handling. Moreover, the ingestion process can be run either manually or set on a recurring schedule, ensuring your metadata stays updated and your tests remain precise and comprehensive.
Importance of Ingesting Schema Metadata
Ingesting metadata into Validatar has several substantial benefits:
Automated Test Building: The metadata information can be used to automatically build tests for your data, making the testing process more efficient.
Data Profiling: Metadata ingestion enables users to perform data profiling, allowing for a comprehensive understanding of the data.
Data Monitoring: It provides an overview of the current state of your data and tracks changes over time, ensuring you're up-to-date on your data's status.
Practical Applications
Metadata ingestion in Validatar is a powerful feature with a broad range of practical applications:
Data Testing: Automatic test building using metadata helps in ensuring and assessing the quality of the data. Automatically generating tests based on the defined metadata ensures that any changes to the data pipeline are promptly validated, reducing the risk of introducing errors.
Schema Profiling: Using Validatar for data profiling needs help data teams report on data distributions, patterns, and anomalies, providing insights into the data quality and helping in making informed decisions about data cleansing or transformation requirements.
Data Monitoring: Ingesting metadata helps data teams monitor changes in key metrics by establishing rules and thresholds based on metadata. Then Validatar can automatically detect and notify the team of any deviations, allowing for proactive intervention.
Data Governance & Compliance: Validatar can help enforce data governance by validating that data conforms to the predefined standards and policies outlined in the metadata. This ensures that data is consistently handled and meets compliance requirements.
Data Lineage Analysis: Validatar, by using metadata, can assist in visualizing data lineage. This helps with understanding the flow of data from source to destination being effectively valuable for identifying dependencies, understanding the impact of changes, and troubleshooting issues in complex data pipelines.
How to Configure Metadata Ingestion
Prerequisite
Before configuring schema metadata ingestion, ensure that the Data Source for which you are setting up schema metadata has already been created.
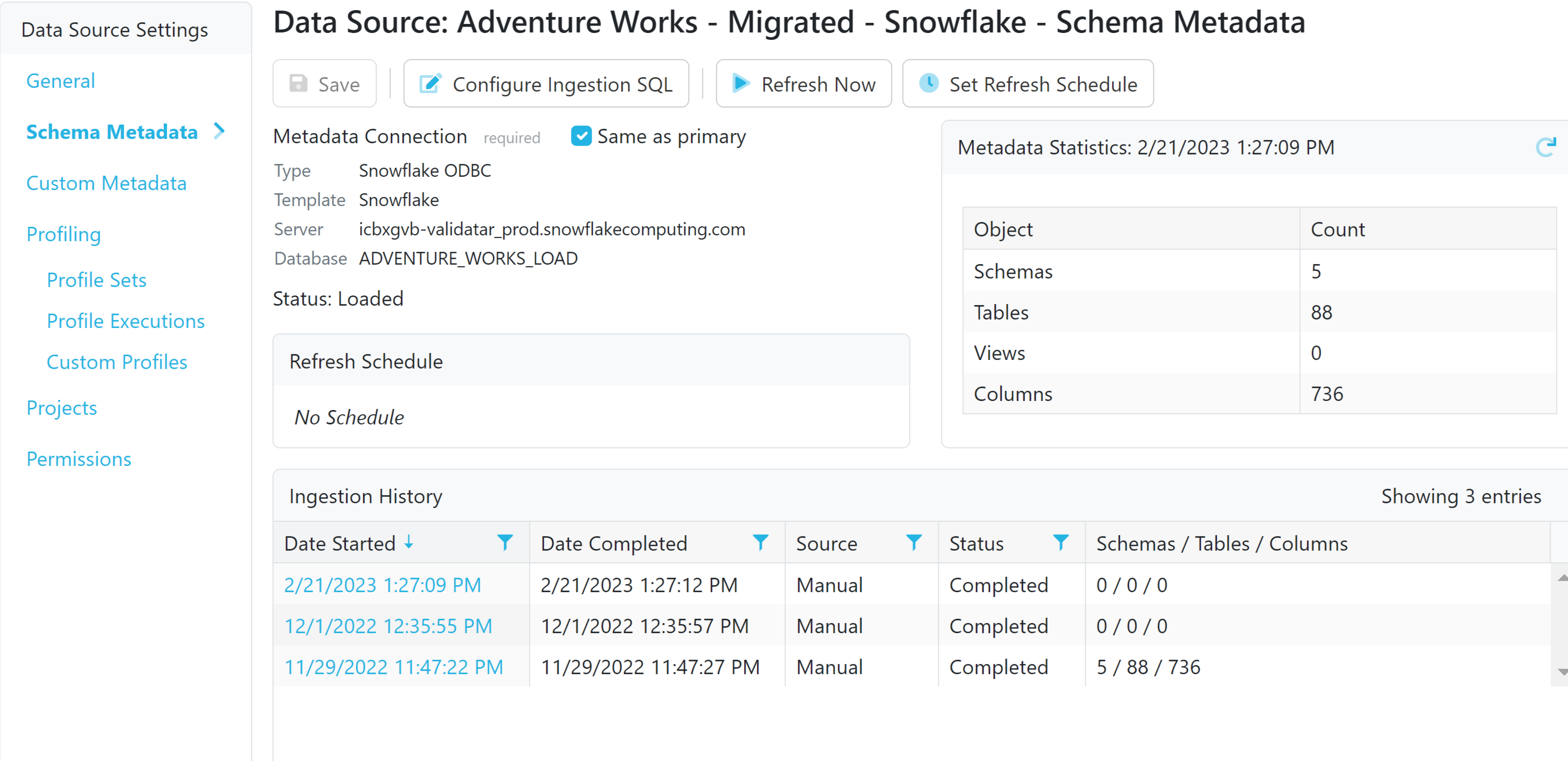
Locate the Data Source
Go to Settings > Data Sources > Select your Data Source.
Within Data Source Settings, choose Schema Metadata.
Configure the Connection
The default metadata connection is the same as the one used for pulling data for test results. Keep this default or add a secondary connection by unchecking 'Same as Primary' and clicking 'Add'. The process mirrors creating a connection for a new data source.
Select Metadata with SQL
Customize SQL scripts for each object type. While SQL scripts are provided by default for SQL Server and Snowflake, you can override and customize them. For Python-based scripts, only one combined script containing each DataFrame variable is required.
Refer to the table in the section below for the required columns for each object type/ dataframe.
Preview and finalize the configuration for each object by reviewing the scripts. Click 'Update Ingestion SQL' to save.
Ingest Metadata
After configuring the SQL, click 'Refresh Now' to extract metadata from your data source and load it into Validatar.
View Results
To view ingested metadata, click on the date and time under Ingestion History. The pop-up displays the runtime schema metadata SQL, along with the list of inserted schemas, tables, and columns. Navigate to specific levels by clicking the object tabs.
Monitoring Changes
Subsequent runs of schema metadata ingestion display lists of inserted, updated, and deleted objects since the last ingestion. Use this information to monitor changes in your metadata.
Writing Metadata Ingestion Scripts
This table displays the necessary columns for the metadata ingestion scripts. Custom metadata fields are supported in the ingestion script and are optional. To simplify the process, all columns are conveniently listed and you can drag-and-drop each column into the Validatar query window.
Object Type / DataFrame | Required Columns | Optional Columns |
|---|---|---|
Schema | Name* | |
Table | Schema*, Name* | Type, ViewDefinition |
Column | Schema*, Table*, Name*, DataType, Sequence | IsNullable, IsIdentity, IsPrimaryKey, StringMaxLength, NumericPrecision, NumericScale, DateTimePrecision, Comment |
* = Column must be unique
Here are examples from Snowflake demonstrating how you can write schema metadata ingestion scripts for each database object.
Schema-Level
select schema_name as "Name",
/* The following line is a Custom Metadata Field example */
last_altered as "LastAltered"
from information_schema.schemata
where schema_name <> 'INFORMATION_SCHEMA';Table-Level
select
t.table_schema as "Schema",
t.table_name as "Name",
case when t.Table_Type = 'VIEW' then 'V' else 'T' end as "Type",
v.View_Definition as "ViewDefinition"
from information_schema.tables t
left join information_schema.views v on t.table_catalog = v.table_catalog and t.table_schema = v.table_schema and t.table_name = v.table_name
where t.table_schema <> 'INFORMATION_SCHEMA';Column-Level
select c.TABLE_SCHEMA as "Schema",
c.TABLE_NAME as "Table",
c.COLUMN_NAME as "Name",
c.DATA_TYPE as "DataType",
c.ORDINAL_POSITION as "Sequence",
case when c.IS_NULLABLE = 'YES' then 1 else 0 end as "IsNullable",
case when c.IS_IDENTITY = 'YES' then 1 else 0 end as "IsIdentity",
0 as "IsPrimaryKey",
c.CHARACTER_MAXIMUM_LENGTH as "StringMaxLength",
c.NUMERIC_PRECISION as "NumericPrecision",
c.NUMERIC_SCALE as "NumericScale",
c.DATETIME_PRECISION as "DateTimePrecision",
c.COMMENT as "Comment"
from information_schema.COLUMNS c
where c.table_schema <> 'INFORMATION_SCHEMA';Python Combined Script
Here is an example of the combined metadata ingestion Python script used for a Windows Directory - Delimited Files data source.
### SETUP DATAFRAMES ###
schema_dataframe = pd.DataFrame(columns=['Name','contains_files','schema_object_represents'])
table_dataframe = pd.DataFrame(columns=['Schema','Name','Type','file_type','date_modified','file_size','file_extension','column_delimiter','file_logical_table'])
column_dataframe = pd.DataFrame(columns=['Schema','Table','Name','DataType','Sequence'])
### HELPER FOR FORMATTING SIZE AS HUMAN READABLE
def sizeof_fmt(num, suffix="B"):
for unit in ["", "Ki", "Mi", "Gi", "Ti", "Pi", "Ei", "Zi"]:
if abs(num) < 1024.0:
return f"{num:3.1f}{unit}{suffix}"
num /= 1024.0
return f"{num:.1f}Yi{suffix}"
### SCHEMA TABLE COLUMN INGESTION SCRIPT ###
folders = [parent_folder_path]
if include_sub_folders:
for root, dirs, files in os.walk(parent_folder_path):
for name in dirs:
folders.append(os.path.join(root, name))
### GET ALL SCHEMAS AND TABLES ###
for folder in folders:
schema_name = folder.replace(parent_folder_path,'.')
schema_dataframe_new = pd.concat([schema_dataframe, pd.DataFrame({'Name':[schema_name],'contains_files':['Yes'],'schema_object_represents':['Folder']})], ignore_index = True)
schema_dataframe_new.reset_index()
schema_dataframe = schema_dataframe_new
file_names = [f for f in os.listdir(folder) if os.path.isfile(os.path.join(folder, f))]
schema_names = [schema_name for f in file_names]
table_type ='Table'
file_type ='Delimited'
file_types = [file_type for f in file_names]
file_sizes = [sizeof_fmt(os.path.getsize(folder + '/' + f)) for f in file_names]
date_modifieds = [datetime.fromtimestamp(os.path.getmtime(folder + '/' + f)).strftime('%Y-%m-%d %H:%M:%S') for f in file_names]
file_extensions = [acceptable_file_extensions for f in file_names]
column_delimiters = [delimiter for f in file_names]
file_logical_tables = [f.split('-')[0].split('.')[0] for f in file_names]
table_type_names = [table_type for f in file_names]
table_dataframe_new = pd.concat([table_dataframe, pd.DataFrame({'Schema':schema_names,'Name':file_names,'Type':table_type_names,'file_type':file_types,'date_modified':date_modifieds,'file_size':file_sizes,'file_extension':file_extensions,'column_delimiter':column_delimiters,'file_logical_table':file_logical_tables})], ignore_index = True)
table_dataframe_new.reset_index()
table_dataframe = table_dataframe_new
### ELIMINATE UNACCEPTABLE FILES FROM TABLES
starting_table_dataframe = pd.DataFrame(columns=['Schema','Name','Type','file_type','date_modified','file_size','file_extension','column_delimiter','file_logical_table'])
for extension in acceptable_file_extensions.split(','):
starting_table_dataframe = pd.concat([starting_table_dataframe,table_dataframe[table_dataframe.Name.str.endswith(extension)]])
table_dataframe = starting_table_dataframe
table_dataframe.reset_index()
###
### LOOP THROUGH ALL FILES AND GET COLUMN INFO
for index, row in table_dataframe.iterrows():
try:
fullpath = parent_folder_path + row['Schema'][1:] + '\\' + row['Name']
if first_row_contains_column_names:
x = pd.read_csv(fullpath, low_memory=False, delimiter=delimiter, header=0, nrows=number_of_records_to_preview)
else:
x = pd.read_csv(fullpath, low_memory=False, delimiter=delimiter, header=None, nrows=number_of_records_to_preview)
column_dataframe_new = pd.concat([column_dataframe, pd.DataFrame({'Schema':row['Schema'],'Table':row['Name'],'Name':x.columns,'DataType':x.dtypes,'Sequence':numpy.arange(len(x.columns))+1})], ignore_index = True)
column_dataframe_new.reset_index()
column_dataframe = column_dataframe_new
except Exception as ex:
print(ex)
Automating Metadata Ingestion
Metadata ingestion can be configured to run on a recurring schedule. To set this up:
Ingestion Settings: Navigate to the Schema Metadata settings of your data source.
Scheduling: Select 'Set Refresh Schedule'.
Defining Schedule: Define the frequency and timing of your recurring ingestion.
Save Settings: Save your settings to start the automated ingestion process. Ensure the schedule is enabled before saving.
Conclusion
Metadata ingestion is a pivotal feature of Validatar that provides benefits in automated test building, data profiling, and data monitoring. It applies to both database-based and script-based data sources and can be run manually or on a recurring schedule. By leveraging the power of metadata ingestion, users can achieve more efficient and comprehensive data management and testing.